Project:Blood typing: Difference between revisions
Mycoplasma (talk | contribs) No edit summary |
Mycoplasma (talk | contribs) |
||
| Line 294: | Line 294: | ||
'''Conclusions:''' | '''Conclusions:''' | ||
Most likely this means that I mixed up chelex and ethanol extractions for the run on the previous day, and chelex actually worked then too. Less likely is that chelex worked this time but not last time, either because I removed PCR inhibitors with the centrifuge, or because we avoided the problem of small reaction volume discussed previously. This would also mean that the ethanol precipitated sample has stopped working. Need to do more to confirm. | Most likely this means that I mixed up chelex and ethanol extractions for the run on the previous day, and chelex actually worked then too. Less likely is that chelex worked this time but not last time, either because I removed PCR inhibitors with the centrifuge, or because we avoided the problem of small reaction volume discussed previously. This would also mean that the ethanol precipitated sample has stopped working. Need to do more to confirm. Odd that chelex genomic showed such a strong band last time but none this time. | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
Revision as of 22:26, 1 November 2012
How genes code for blood group
Blood group is determined by the combination of A and B antigens in your red blood cells. A and B individuals have only their respective corresponding antigens, AB individuals have both, and O individuals have none.
The production of these antigens is determined by the "histo-blood group ABO system transferase" gene (1062 base pairs), which is part of the ABO gene locus. This gene codes for the expression of a glycosyltransferase enzyme which by acting on another antigen(H), produces A or B antigens. The A and B allelic forms of the gene code for different forms of glycosyltransferase which affect the H antigen in different ways. O alleles code for another protein that doesn't affect the H antigen, meaning no A or B antigens are expressed. O alleles have a deletion of G at 258, while B alleles have a single nucleotide polymorphism (SNP) from G to A at position 700. The gene we're looking at is here: ABO gene. You need to set the origin to 29 under the tools menu, as this viewer is for the entire ABO gene; the histo-blood group ABO system transferase gene doesn't start until position 29.
Each person has two of these alleles, one from each parent. A and B are dominant, O is recessive, so the possible combinations are:
| Alleles | Blood group |
|---|---|
| AA | A |
| AO | A |
| BB | B |
| BO | B |
| AB | AB |
| OO | O |
In the UK the distribution of A, B, AB and O is 42%, 10%, 4% and 44%.
Process overview
1) Obtain two sequences of DNA through PCR, the first containing the deletion site at 258, and the second containing the SNP site at 700
2) Use restriction enzyme KpnI on the first fragment to cut the O alleles only, and use restriction enzyme AluI on the second fragment to cut the B alleles only. (KpnI's cutting site is GGTAC^C - this sequence is found in 0 alleles, but in A and B alleles the sequence is GGTGACC, hence they are not cut. AluI's cutting site is AG^CT - in A and O alleles the initial A is a G, hence only B alleles are cut.)
3) Do gel electrophoresis on the resulting fragments, hopefully resulting in distinguishable bands to show the 6 different alleles. We should then be able to determine an individual's blood type.
Process reality
Much of the necessary equipment we already have from the sex typing experiments. Of the new things, we need the restriction enzymes, and possible a new gel - see below.
All fragments in the papers are between 80 and 200 bp long. For this we would need a polyacrylamide gel (which we have decided against due to difficulty with handling) or a high quality agarose concentrated at 3%, which is a bit expensive, but not impossible. So we have decided to design our own primers to get larger fragments to work with.
Design of primers for larger fragments:
Using the ApE software, we have found a set of primers to enable us to use longer fragments. Here is the file with the primer sequences (Save this link as a file): ApE file, and Download ApE here
Primers for G deletion sequence:
| Primer | Sequence | Length | GC% | Tm(°C) |
|---|---|---|---|---|
| P1 forward | 17484 5' CCCGCAGGTCCAATGTTGAG 3' 17503 | 20 | 60 | 59 |
| P1 reverse | 18268 5' ATCTGACAGAGAAGTGACCACG 3' 18247 | 22 | 50 | 58 |
Product is 784bp.
After digestion with KpnI you get two sizes of fragments - 243bp and 541bp
Primers for G to A SNP:
| Primer | Sequence | Length | GC% | Tm(°C) |
|---|---|---|---|---|
| P2 forward | 19125 5' GAGGTGGATTACCTGGTGTGC 3' 19145 | 21 | 57 | 59 |
| P2 reverse | 19473 5' GCACCTTGGTGGGTTTGTGG 3' 19454 | 20 | 60 | 60 |
Product: 347 bp.
After digestion with AluI : two fragment sizes of 96 bp and 251 bp
New equipment
Sigma aldrich have KpnI and AluI. I couldn't find them on NBS bio. A cheaper alternative is to try to get them from NEB (http://www.neb.uk.com/).
If we do need high quality agarose here is sigma's selection. NBSbio have agarose but no info on its purity.
Sources
Most of the procedure came from this paper. With some more here. For other papers and background see here.
Here is a sequence viewer for the ABO gene. Histo-blood group ABO system transferase starts at position 28 - so to find the deletion at 258 and the SNP at 700 you have to add 28 to the numbers on the viewer. Go from the 5' end.
Extraction and PCR 15/8
- Extract DNA (two samples, Will and Nicholas) from cheek by rubbing with pipette tip and taking saliva
- Mix with 250ul of chelex 100, cover with a drop of oil and incubate at 56°C for 30 mins.
- Centrifuge for 5 mins and remove supernatant. Supernatant kept to view genomic DNA later.
- Two reactions prepared - Nicolas (AMELX/Y primers), Will (PB1 primers)
- Reaction volume - 12.5ul Taq, 5ul template, 5ul primers, 2.5ul deionised water.
- PCR: initial denaturation at 96°C for 5 mins, then 35 cycles of 96°C for 1 min, annealing at 55° for 30 secs, extension at 72°C for 1 min.
- Results. PCR failed. Will's DNA was the one using blood typing primers.
Extraction and electrophoresis of genomic DNA 15-17/8
- Add cheek scraping + saliva to 1/2 a PCR tube of chelex (half bead layer, half liquid layer). Mix
- Incubate at 56°C for 30 mins, vortex briefly, 94°C for 10 mins. (Should have centrifuged and) take supernatant.
- Add 5ul of loading buffer to 25ul of template - visualise on 1% agarose gel
- DNA visible in well.
Extraction and PCR 19-20/8/12 (Will)
- Chelex extraction. Visualised and saw glow in well so proceeded to use this as template.
- 25ul total reaction volumes. Quantities assumed as no record: 5ul template (1ul for PC), 5ul primers, 12.5ul Taq readymix, 2.5ul dH20 (6.5ul for PC).
- Initial D - 96C 5 mins, then 35 cycles of D-96C 1 min, A-55C 30 secs, E-72C 1 min.
From right:
- 1: 250-10000bp ladder (from hackspace). Ladder key
- 2: PC + amelogenin (sex typing) primers (combined aliquot)
- 3: PC + amelogenin (sex typing) primers (separate aliquots)
- 4: PC + PB1 primers
- 5: Template put through PCR program with no other reagents
- 6: Template + separate AMEL primers
- 7: Template + separate PB1 primers
- 8: Template - no PCR
Conclusions:
- As PC worked with PB1 primers but not AMEL, there is either a problem with the AMEL primers, or the annealing temperature of 55C doesn't work for them.
- Something in the reagents / template stops PCR working for our extractions OR there is too little DNA extracted. However even if the extracted DNA is very little, surely is enough for PCR? And we are pretty sure there is DNA there because it shows up genomically in the wells.
Extraction and PCR 03/10
- About 2ul of saliva scraped from cheek + 325ul chelex (half bead layer, half liquid layer). Mix.
- Incubate at 56°C for 30 mins (missed vortex), 94°C for 10 mins. Centrifuge and take supernatant.
- PCR mix: 5ul template, 5ul primers, 2.5ul dH20, 12.5ul Jumpstart taq readymix
- Positive control: 1ul template, 5ul primers, 6.5ul dH20, 12.5ul Jumpstart taq readymix
- PCR: 94C 2min, 30 cycles (94C 30s, 56C 30s, 72C 2min), 72C 5min.
From left:
- 1: 250-10000bp ladder (from hackspace). Ladder key
- 2: Positive control PCR product.
- 3: Mike PCR product
- 4: Simon PCR product
- 5: Will PCR product
- 6: MF1 (From UCL)
- 7: N.H / N.1+ (From UCL)
The positive control worked (You can see if you look closely). However the band was significantly weaker than the gel run on August 20th with same PC + same primers. None of the PCRs on our extractions worked.
I also ran our 3 genomic samples, and nothing was there - providing a reason for PCR failure.
Extraction 25/10 (Done by Will and Lui)
- 10ml dH20 vigorously rinsed in mouth (with cheek biting) for 3 mins
- Spat out, added ∼1.5ml washing up liquid, mixed, left for ∼10 mins.
- Split samples into two groups
1st group:
- Took ∼500ul sample, added ∼100ul Proteinase K solution
- Incubated for 1hr at 50C, 10 mins 94C
- Added 1 volume 100% EtOh, small amount of precipitate observed, so centrifuged at 13000 rpm for 20 mins
- Dried at ∼50C
- Added ∼350ul 70% IPA, centrifuged at 13000 rpm for 10 mins
- Air dried pellet
- Tried to resuspend in 500ul TE but pellet wouldn't resuspend completely. Unsure if none had dissolved or some had. Even incubating at 60C didn't work. Edit - seemed none dissolved as nothing showed up on gel. Didn't dissolve even in 10 ml dH20.
2nd group (remainder of sample - 11ml:
- Added 10 shakes meat tenderiser
- Added 1/2 volume 70% IPA. Significant amount of precipitate observed. Able to remove it with pipette.
- Dried at ∼50C
- Split, washed one with 70% IPA, centrifuged 10min at 13000 RPM. Then resuspended pellet in 500ul TE. Other only resuspended, no wash. Resuspensions didn't totally work (not sure if any dissolved).
PCR of 1st group 27/10 (Will's sample)
- 25ul total volume - 5ul template (1ul for PC), 5ul primers, 12.5ul Taq readymix, 2.5ul dH20 (6.5ul for PC)
- PCR was Initial D - 95C 5 mins, then 35 cycles of D-96C 1 min, A-55C 30 secs, E-72C 1 min.
- Gel - loaded 5ul of ladder, 15ul of each sample, ∼ 0.2 vol loading buffer
From right (after 35 mins at ∼ 80V:)
- 1: 250-10000bp ladder. Ladder key (At the far right, you can barely see it)
- 2: Will PCR product (PB1 primers) (Nothing)
- 3: Positive control PCR product (PB1 primers) - We expect this to be 784 bp, looks right.
- 4: Positive control PCR product (PB2 primers) - We expect this to be 347 bp, looks right.
- 5: Will genomic (Nothing)
Conclusions:
- Something went wrong with the ladder - it's barely visible. I used standard quantity so unsure why.
- My extractions failed PCR, and nothing showed up in genomic - hence extractions failed. If the pellet was DNA and had not degraded, it just must not have dissolved.
- Both controls worked - so both sets of our blood typing primers work.
Extraction 28/10
- Used this protocol. No DNA precipitated, so resolved to try again tomorrow using cheek swish method instead of cheek scrape.
Extraction 29/10
- Used this protocol. Instead of scraping cells from cheek, swished 5ml of dH20 in mouth for 3 mins while chewing cheeks. Then took 200ul of this and proceeded with extraction.
- One reaction had 1%Triton. 150ul of sample + lysis buffer added to 50ul PK. Incubated. No precipitate observed, but washed where pellet would be with 70% Etoh. Stored in fridge in 70%Etoh.
- One reaction had 10%Triton. 150ul of sample + lysis buffer added to 50ul PK. Incubated. No precipitate observed, but washed where pellet would be with 70% Etoh. Stored in fridge in 70%Etoh.
- Added 0.5ml dishwashing detergent to 'mouth swish remainder' (4.5ml). Added a few shakes meat tenderiser. Took ∼150ul, centrifuged and took supernatant. Added 0.25 vol PK and incubated. Precipitated and washed, small amount of DNA observed, tried to resuspend but wouldn't dissolve. Stored at -20C.
- Used IPA to precipitate the rest. Large pellet extracted, air dryed for 5 mins, then would not dissolve in 300ul TE at 65C. Stored at -20C.
Extraction and PCR 31/10 (Will)
- Extracted DNA from cheek using this protocol
- Ran PCR with PB2 primers on this sample + the third sample from the 29/10 + a positive control.
- 25ul total volume - 5ul template (1ul for PC), 5ul primers, 12.5ul Taq readymix, 2.5ul dH20 (6.5ul for PC)
- Initial D - 95C 5 mins, then 35 cycles of D-96C 1 min, A-55C 30 secs, E-72C 1 min.
From left (after ∼40 mins at 80V:)
- 1: 250-10000bp ladder. Ladder key
- 2: Will PCR product (Chelex extraction) (Nothing there)
- 3: Will PCR product (Ethanol extraction) - We expect this to be 347 bp, looks right.
- 4: Positive control PCR product - We expect this to be 347 bp, looks right.
- 5: Chelex extraction genomic
- 6: Ethanol extraction genomic (Nothing there)
Conclusions:
This needs to be repeated to confirm, but it seems to show that the ethanol precipitated DNA worked in PCR, even though it looked like none had dissolved, while the chelex didn't. If at least one has worked that's good news. It's strange though that the chelex extraction shows up as genomic in the well, but failed PCR, whereas the ethanol extraction is the other way round. Possible that something in chelex mix inhibited PCR, while quantity of genomic was too little to show up for ethanol, but was enough for PCR. nb. Chelex PCR product was oddly small compared to other.
PCR 1st Nox 2012 (Will)
Repeated PCR from 31 Oct to test conclusions.
- Same inputs except that the chelex solution was centrifuged and resulting pellet discarded before PCR. Also another sample from 29/10 used in addition to one from last time. PB2 primers used.
- 25ul intended total volume - 5ul template (1ul for PC), 5ul primers, 12.5ul Taq readymix, 2.5ul dH20 (6.5ul for PC). Actually put 6.5ul of dH20 in each sample by mistake, meaning total volume of 29ul for all except PC.
- Initial D - 95C 5 mins, then 35 cycles of D-96C 1 min, A-55C 30 secs, E-72C 1 min.
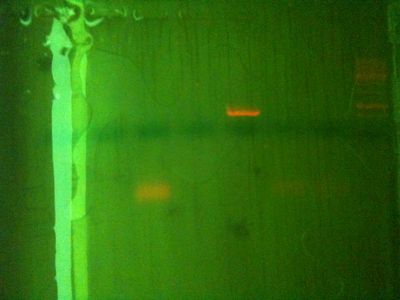
From left (after 40 mins at 80V:)
- 1: 250-10000bp ladder. Ladder key
- 2: Will PCR product (Chelex extraction) - We expect this to be 347 bp.
- 3: Will PCR product (3rd Ethanol extraction from 29/10) - Nothing
- 4: Will PCR product (1st Ethanol extraction from 29/10) - Nothing
- 5: Positive control PCR product - We expect this to be 347 bp
- 6: Chelex extraction genomic
Lanes 7 and 8 not visible on photo
- 7: 3rd Ethanol extraction genomic (Nothing there)
- 8: 1st Ethanol extraction genomic (Nothing there)
Conclusions:
Most likely this means that I mixed up chelex and ethanol extractions for the run on the previous day, and chelex actually worked then too. Less likely is that chelex worked this time but not last time, either because I removed PCR inhibitors with the centrifuge, or because we avoided the problem of small reaction volume discussed previously. This would also mean that the ethanol precipitated sample has stopped working. Need to do more to confirm. Odd that chelex genomic showed such a strong band last time but none this time.