Difference between revisions of "Project:JuicyPrint"
(Created page with "=Overview= ==DIYbio== * London Biohackspace is a community biology lab based at the London Hackspace in Hackney, London. * As members of the DIYbio community we enjoy creating...") |
|||
| Line 1: | Line 1: | ||
=Overview= | =Overview= | ||
==DIYbio== | ==DIYbio== | ||
| − | * | + | * We hope JuicyPrint will be possible to construct and operate with only "off-the-shelf" items and our strain of bacteria. |
| − | |||
| − | |||
| − | |||
* G. hansenii (Gluconacetobacter hansenii) is able to grow on a wide range of carbon sources such as fruit juice, brewing waste or tea. So once a printer unit is constructed, creating objects from it will require only a computer and a local grocery shop. | * G. hansenii (Gluconacetobacter hansenii) is able to grow on a wide range of carbon sources such as fruit juice, brewing waste or tea. So once a printer unit is constructed, creating objects from it will require only a computer and a local grocery shop. | ||
| Line 128: | Line 125: | ||
'''c-di-GMP:''' known to regulate biofilm formation, motility, and the production of extracellular polysaccharides. c-di-GMP acts as a positive regulator of cellulose synthase in ''G. hansenii'', thus a positive regulator of extracellular cellulose production. | '''c-di-GMP:''' known to regulate biofilm formation, motility, and the production of extracellular polysaccharides. c-di-GMP acts as a positive regulator of cellulose synthase in ''G. hansenii'', thus a positive regulator of extracellular cellulose production. | ||
| + | |||
| + | [[Category:Biohacking]] | ||
Revision as of 12:05, 14 September 2014
Overview
DIYbio
- We hope JuicyPrint will be possible to construct and operate with only "off-the-shelf" items and our strain of bacteria.
- G. hansenii (Gluconacetobacter hansenii) is able to grow on a wide range of carbon sources such as fruit juice, brewing waste or tea. So once a printer unit is constructed, creating objects from it will require only a computer and a local grocery shop.
G. hansenii
- Gluconacetobacter hansenii (G. hansenii) is a species of acetic acid bacteria, notable as the model organism for the biosynthesis of bacterial cellulose.
- G. hansenii used to be lumped in with Acetobacter xylinum before being reclassified as a seperate species.
- Our work is centred around strain LMG 1524 (ATCC 23769) for which an entire genome sequence is available.
- It can be found as part of the community naturally found in fermenting kombucha tea, though the strain we are using was originally isolated from vinegar.
Engineering light sensing G. hansenii
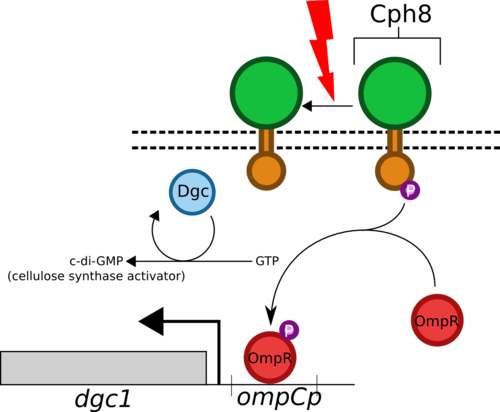
- The Cph8 biobrick encodes for a fusion of EnvZ (osmolarity sensing histidine kinase from E. coli) and Cph1 (photoreceptor from cyanobacteria Synechocystis).
- Cph8 phosphorylates OmpR under normal condiitions but when exposed to red light (660nm) the phosphorylation is inhibited.
- OmpR forms a homodimer when phosphorylated and binds to the ompC promoter region (also a biobrick) enhancing the transcription of whatever gene follows it.
- We will place dgc1, which encodes for Dgc (diguanylate cyclase), under the refulation of this promoter.
- Dgc catalyses formation of c-di-GMP in G. hansenii, which then activates cellulose synthase and begins the production of extracellular cellulose.
- We will produce a dgc1 knockout strain of G. hansenii so that all dgc1 transcription will be controlled by our pathway.
- Shining red light on our strain will therefore block this pathway and prevent cellulose production.
Biobricks
- OmpR is the missing piece in the biobrick repository for using the light Cph8 light sensing pathway in organisms other than E. coli.
- We are producing an OmpR biobrick so that we can work with the pathway in G. hansenii but it will hopefully also enable its use in wider range of organisms.
- This also involves correcting the currently mislabelled parts in the iGEM repository - parts which contain the ompC promoter are currently labelled as "OmpR".
- We are also hoping to produce a biobrick which contains all the individual parts to test whether the Cph8 pathway can be used in an organism of choice simply by adding a constitutive promoter from that organism.
Hardware
- Gluconacetobacter normally produces a flat sheet of cellulose across the surface of the liquid medium it is grown in.
- Our strain of G. hansenii will not produce cellulose when red light is shone onto it.
- By shining a pattern of red light onto the tub of culture, a layer of cellulose can be produced which is the negative of the red light pattern.
- Pushing this layer just below the surface of the medium will allow a new layer, with a new pattern, to grow on top of this layer and fuse with the previous layer.
- Repeating this process allows a 3D form to be built up out of differently shaped layers.
Further Uses
- A 3-dimensional structure of hydrated cellulose has similar properties to collagen and offers a great opportunity for creating tissue scaffolds for use in regenerative medicine.
- Dried bacterial cellulose has a very high tensile strength and has been used in areas from fashion design to medical wound dressings.
- Being able to print a 3D structure offers the possibility of creating textiles with novel properties and even entire objects from vases to structural components of devices.
Details of Relevant Genes
The genes in the table below are all the genes relevant to our proposed light sensing system to be inserted into G. hansenii.
Genes highlighted in pink are genes that will actually be used in our system.
| Gene | Function | Description | Origin | Biobrick |
|---|---|---|---|---|
| envZ | Encodes EnvZ | EnvZ is a histidine kinase/phosphatase found in E. coli. EnvZ responds to the osmolarity changes in the medium through variations in membrane surface tension triggering conformational changes and phosphorylates/dephosphorylates OmpR. Low osmolarity -> dephosphorylation of OmpR. High osmolarity -> phosphorylation of OmpR. | E. coli | |
| cph1 | Encodes Cph1 | Cph1 is a cyanobacterial phytochrome (protein in which exposure to light induces conformational changes). | Synechocystis | |
| cph8 | Encodes Cph8 | Cph8 is a fusion of the light responsive domain of Cph1 and the histidine kinase domain of EnvZ. The light responsive domain (Cph1) has maximal response to light near 660nm. Exposure to red light inhibits the activity of the EnvZ histidine kinase domain. When active (in the dark) its kinase domain phosphorylates endogenous OmpR. Light sensitivity is only functional in the presence of Phycocyanobilin (PCB). | Engineered | BBa_I15010 or BBa_K1017301 |
| ho1 | Encodes Ho1 | Heme oxygenase (Ho1) is a product of ho1. One of two proteins required for the biosynthesis of PCB from heme, thus is required to produce functional Cph1 or Cph8. | Synechocystis | BBa_I15008 |
| pcyA | Encodes PcyA | Phycocyanobilin:ferredoxin oxidoreductase (PcyA) is the second of two proteins required for the biosynthesis of PCB from heme. Thus it is required to produce functional Cph1 or Cph8. | Synechocystis | BBa_I15009 |
| ompR | Encodes OmpR | OmpR is a DNA-binding protein (transcription factor) that is phosphorylated or dephosphorylated by EnvZ depending the osmolarity of the medium. Phosphorylated OmpR (OmpR-P) binds to sites within ompCp and ompFp. Low levels of OmpR-P upregulate ompF transcription. High levels of OmpR-P upregulate ompC transcription and repress ompF transcription (see Egger et al., 1997). BEWARE: The iGEM repository uses ompR and OmpR where they mean to say ompCp/OmpR-binding site. |
E. coli | (BBa_K098011) |
| ompC | Encodes OmpC | OmpC is an outer membrane porin found in E. coli. | E. coli | |
| ompCp | Regulates ompC transcription | ompCp is the upstream operator-promoter region of ompC. ompCp contains three OmpR-P binding sites: the C1, C2, and C3 sites located in the the -100 to -38 region of ompC. Higher levels of OmpR-P lead to increased rate of binding and lead to upregulation of ompC transcription. | E. coli | BBa_R0082 |
| dgc1 | Encodes Dgc | Diguanylate cyclase (Dgc) catalyses the formation of c-di-GMP in G. hansenii. Dgc is necessary for extracellular cellulose. Dgc is a product of dgc1 (possibly also produced by dgc2 but despite dgc2’s sequence homology with dgc1, dgc2 cannot substitute for the function of dgc1 under normal growth conditions). | G. hansenii | |
| ompF | Encodes OmpF | OmpF is an outer membrane porin found in E. coli. | E. coli | |
| ompFp | Regulates ompF transcription | ompFp is the upstream operator-promoter region of ompF. ompFp contains four OmpR-P binding sites: the -380 to the -361 region (F4 site), and the -100 to -39 region (F1, F2, and F3 sites) of ompF. With low levels of OmpR-P only the F1, F2, and F3 sites are bound leading to upregulated transcription of ompF. With high levels of OmpR-P the F4 site is also bound and leads to repression of ompF transcription. | E. coli |
Additional terms:
ompB: E. coli operon containing ompR and envZ which produce the gene products OmpR and EnvZ respectively. ompR and envZ are transcribed together as a polycistronic mRNA and regulated in a complex fashion by the interaction between the ompB operator-promoter region and cyclic AMP-CRP.
cdg1: The first cdg (cyclic diguanylate) operon of the two homologous operons (cdg1 and cdg2) in G. hansenii. cdg1 contains dgc1.
c-di-GMP: known to regulate biofilm formation, motility, and the production of extracellular polysaccharides. c-di-GMP acts as a positive regulator of cellulose synthase in G. hansenii, thus a positive regulator of extracellular cellulose production.