Difference between revisions of "Project:Public Biobrick"
Pingutastic (talk | contribs) |
m (Montyphy moved page Public Biobrick to Project:Public Biobrick) |
||
| (36 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
== Overview == | == Overview == | ||
| − | UCL's [http://2012.igem.org/Team:University_College_London iGem team] got in touch with us in May about a collaboration with their project to make a [http://partsregistry.org/Help:An_Introduction_to_BioBricks biobrick] that could help to clean up plastic floating in the ocean. From August the biohackers have been working with them to develop a 'public biobrick' (ie. one developed outside of the traditional university or research lab). This biobrick contains antifreeze | + | UCL's [http://2012.igem.org/Team:University_College_London iGem team] got in touch with us in May about a collaboration with their synthetic biology project to make a [http://partsregistry.org/Help:An_Introduction_to_BioBricks biobrick] that could help to clean up plastic floating in the ocean. From August the biohackers have been working with them to develop a 'public biobrick' (ie. one developed outside of the traditional university or research lab). This biobrick contains antifreeze from the marine bacteria Oceanibulbus Indolifex in a plasmid backbone called [http://partsregistry.org/Part:pSB1C3 PSB1C3 ] which contains choloramphenical resistance gene. |
| + | |||
| + | The antifreeze gene when put into bacteria would allow them to gain frost resistance. Antifreeze proteins bind to small ice crystals to inhibit growth and recrystallization of ice that would otherwise be fatal. | ||
| + | |||
| + | Here is the UCL [http://wiki.london.hackspace.org.uk/w/images/a/af/DIYbio_Collaboration_protocols.pdf document] listing all the protocols we used. | ||
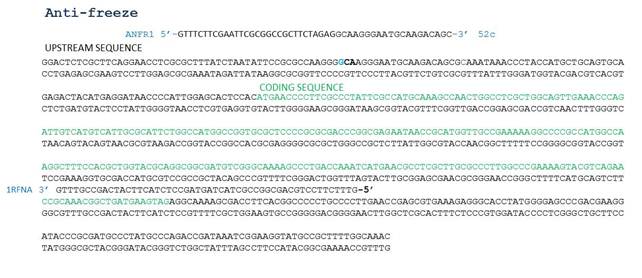
=== Designing primers to amplify the antifreeze and mercuric reductase genes from O. Indolifex === | === Designing primers to amplify the antifreeze and mercuric reductase genes from O. Indolifex === | ||
| + | Primers designed by Nicholas | ||
| + | |||
| + | [http://www.ebi.ac.uk/ena/data/view/EDQ05862 Antifreeze Sequence] | ||
| + | |||
| + | [[File:Primer ANF.jpg]] | ||
| − | + | Mercuric Reductase Sequence | |
| + | |||
| + | [[File:MR primer.jpg]] | ||
=== Generating competent cells (Hackspace) === | === Generating competent cells (Hackspace) === | ||
| Line 16: | Line 27: | ||
=== DNA extraction from O. Indolifex and PCR of two genes === | === DNA extraction from O. Indolifex and PCR of two genes === | ||
| + | Method | ||
| + | Genomic extraction | ||
| + | *Add 1.5 vol of 100% ethanol + 0.1 volume 3M sodium acetate to 400ul liquid overnight culture of bacteria | ||
| + | *Vortex for 5 s | ||
| + | *Freeze at -80C for 1hr (or -30C overnight done at Hackspace) | ||
| + | *Centrifuge at 12000 rpm for 20 mins (centrifuged at the max speed at Hackspace with their centrifudge) | ||
| + | *Add 1ml of 70% ethanol | ||
| + | *Centrifuge at 12000 rpm for 10 mins , dry pellet | ||
| + | *Resuspended in 30ul of dH2O | ||
| + | |||
==== 3-5/9: UCL ==== | ==== 3-5/9: UCL ==== | ||
| Line 71: | Line 92: | ||
| + | |||
| + | |||
| + | [[File:Genomic extraction.jpg]] | ||
| + | [[File:Genomic extraction 2.jpg]] | ||
==== 14-15/9: Hackspace ==== | ==== 14-15/9: Hackspace ==== | ||
| Line 116: | Line 141: | ||
So we got no bands from the O. indofilex gDNA reactions, but we did get bands at the right position (285 bp) from 2/3 of the ANF template ones. ANF templates were ANF genes that had been previously PCRed with the non prefix/suffix primers. We also got a band from the positive control at the right position (2070bp). | So we got no bands from the O. indofilex gDNA reactions, but we did get bands at the right position (285 bp) from 2/3 of the ANF template ones. ANF templates were ANF genes that had been previously PCRed with the non prefix/suffix primers. We also got a band from the positive control at the right position (2070bp). | ||
| + | |||
| + | [[File:ANF PCR UCL.jpg]] | ||
=== Restriction digest and ligation === | === Restriction digest and ligation === | ||
| Line 184: | Line 211: | ||
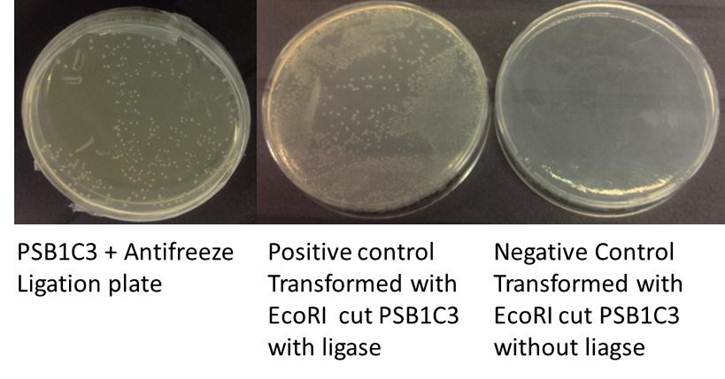
=== Transformation into ''E.coli'' W3110 cells === | === Transformation into ''E.coli'' W3110 cells === | ||
| − | This step was done at UCL laboratory. | + | <div class=""> This step was done at UCL laboratory.</div> |
| + | [[File:Ligation_plate_2.jpg]] | ||
=== Miniprep: Plasmid DNA Extraction === | === Miniprep: Plasmid DNA Extraction === | ||
TBA | TBA | ||
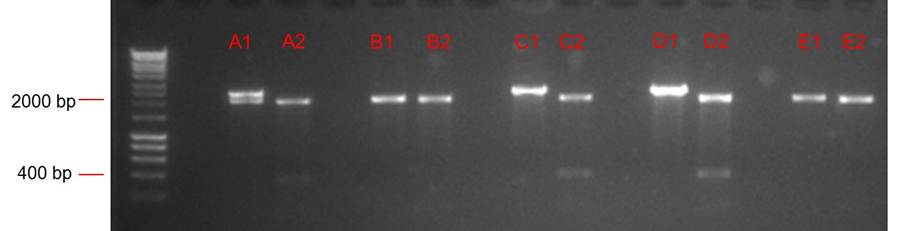
| + | === Analytical digest === | ||
| + | [[File:Analytical digest public biobrick.jpg]] | ||
| + | === Characterisation === | ||
| + | Unfortunately, there was no time to characterize (show it work) the biobrick. There is a simple plate assay that could have been used to test the activity of the protein produced. | ||
| + | Protocol given by UCL: | ||
| + | In order to express the gene, the biobrick we have now need to be cut with restriction enzymes whose restriction sites flank the genes to be ligated with a promoter and RBS (Ribosome binding site). It could be characterized by ligating (sticking) onto an expression vector (a plasmid that is commercially available that contains a promoter and ribosome binding site). | ||
| + | After transformation do the following: | ||
| + | *Pick colony from ligation plates into 2ml LB. Incubate @37 until the OD reach approx 0.5. | ||
| + | *Perform serial dilutions of half of the liquid culture and plate | ||
| + | *Freeze the other half of the culture at -30C overnight. | ||
| + | *Next morning, perform the same serial dilutions on the remaining liquid culture and plate. | ||
| + | *Count the number of colonies on the plate before freezing and after freezing. | ||
| + | |||
| + | [[Category:Biohacking]] | ||
| + | [[Category:Projects]] | ||
Latest revision as of 15:19, 3 June 2013
Overview
UCL's iGem team got in touch with us in May about a collaboration with their synthetic biology project to make a biobrick that could help to clean up plastic floating in the ocean. From August the biohackers have been working with them to develop a 'public biobrick' (ie. one developed outside of the traditional university or research lab). This biobrick contains antifreeze from the marine bacteria Oceanibulbus Indolifex in a plasmid backbone called PSB1C3 which contains choloramphenical resistance gene.
The antifreeze gene when put into bacteria would allow them to gain frost resistance. Antifreeze proteins bind to small ice crystals to inhibit growth and recrystallization of ice that would otherwise be fatal.
Here is the UCL document listing all the protocols we used.
Designing primers to amplify the antifreeze and mercuric reductase genes from O. Indolifex
Primers designed by Nicholas
Mercuric Reductase Sequence
Generating competent cells (Hackspace)
The competent cells are what will take up and express the biobrick at the end of the process. The procedure we followed is here. We built an incubator with shaker to achieve this. After 24 hours we did not see the expected colonies on the plates, we think this is because the streaks were cut too deeply into the growth medium. As an alternative we proceeded with some already prepared plates from UCL that did have colonies.
Amplification of plasmid backbone (Hackspace)
The plasmid backbone is the vehicle for the biobrick, and is the means by which it will get into the competent cells. It is a linearised plasmid backbone containing chlorophenicol resistance. The procedure is in the same link as for competent cells. We did two versions of this reaction, one with the mastermix supplied by UCL, and one with the taq readymix that we use at the hackspace. We saw a band for the readymix one, but not for the mastermix one, so proceeded with the readymix one.
DNA extraction from O. Indolifex and PCR of two genes
Method Genomic extraction
- Add 1.5 vol of 100% ethanol + 0.1 volume 3M sodium acetate to 400ul liquid overnight culture of bacteria
- Vortex for 5 s
- Freeze at -80C for 1hr (or -30C overnight done at Hackspace)
- Centrifuge at 12000 rpm for 20 mins (centrifuged at the max speed at Hackspace with their centrifudge)
- Add 1ml of 70% ethanol
- Centrifuge at 12000 rpm for 10 mins , dry pellet
- Resuspended in 30ul of dH2O
3-5/9: UCL
In this step we attempted to isolate the two genes to be used in the biobrick. The protocol is [TBA here]. Our PCRs failed, and it turned out that no DNA had been extracted by the Generation Column Capture Kit we used. So we proceeded with a previous extraction that had been done with ethanol.
12-13/9: Hackspace
PCR of O. Indoliflex antifreeze gene template provided by UCL with antifreeze primers including biobrick prefix and suffix.
We PCRed the above + a positive control of the plasmid backbone we had previously amplified successfully at the hackspace. No bands were visible on the gel, either for the sample or the positive control, indicating PCR failed. Two theories for why this happened:
- We forgot to add oil to the reactions, running the risk of the reaction mix evaporating. On the other hand not much seemed to have evaporated by the end of PCR.
- We used a PCR program designed for Phusion DNA polymerase and a different type of thermocycler, whereas we used our own thermocycler and our standard taq readymix. The major difference is that we usually have a D step of 30 sec, whereas the UCL protocol calls for a D step of 10s. We also could not do the 10 min final extension step. Finally their ramping speed is about twice ours. Other parameters are similar.
- INFO TO COME - which PCR program we used for the previous successful amplification of PSB1C3 at the hackspace?
Procedure:
- ANF template reaction mix: 2ul of template, 2.5ul FP (ANFR1), 2.5ul RP (1RFNA), 25ul of taq readymix, 18ul dH20
- Positive control PSB1C3 (plasmid backbone) reaction mix: 3ul DNA template, 1ul FP, 1ul RP, 25ul of taq readymix, 20ul dH20
- Initial D - 95C for 30s. Then 30 cycles of (D - 95C for 10s, A - 55C for 25s, E - 72C for 1.5 min). Final extension step of 10min not done. Samples stored at -20C on PCR completion.
Genomic extraction of O. Indoliflex with ethanol.
Both extractions showed up as faint bands in the wells, indicating gDNA extraction succeeded. Bands were same strength as I had previously seen for gDNA extractions at the hackspace, but should have been stronger. This could either be due to the extraction method not extracting enough DNA, or our visualisation technique being inadequate.
Procedure:
- gDNA extraction from the attachment to this email, without the phenol/chloroform steps. Of the options in the protocol, we froze at -20C overnight, and extended the centrifuge steps by 10 mins each due to our less powerful centrifuge. Another difference was that after washing with 70% EtOh, pellet was large and off-white, indicating cellular debris. See photo. So resuspended pellet in 30ul dH20, and discarded everything that did not dissolve. Procedure had previously worked at UCL.
Lanes from right:
- 1: Ladder. Note that loading dye is in the middle of ladder. Possibly because current turned off for 5 mins during gel run?
- 2: ANF
- 3: Positive control PSB1C3
- 4: O. Indofilex gDNA sample 1 (band just visible)
- 5: O. Indofilex gDNA sample 1 (band just visible)
14-15/9: Hackspace
Yeping, Simon and Tonderai prepared and PCRed the following 25ul volume reactions on the 14/9, Will visualised them on the 15/9.
Reagents:
- O. indoliflex gDNA (H): 12.5ul Taq readymix, 2.5ul FP, 2.5ul RP, 1ul gDNA (1:100 concentration), 6.5ul dH20.
- O. indoliflex gDNA (UCL): 12.5ul Taq readymix, 2.5ul FP, 2.5ul RP, 1ul gDNA (1:100 concentration), 6.5ul dH20.
- ANF template 1 from O. indoliflex: 12.5ul Taq readymix, 2.5ul FP, 2.5ul RP, 2ul DNA, 5.5ul dH20.
- ANF template 2 from O. indoliflex: 12.5ul Taq readymix, 2.5ul FP, 2.5ul RP (primers diluted 1:2), 2ul DNA, 5.5ul dH20.
- ANF template 3 from O. indoliflex: 12.5ul Taq readymix, 2.5ul FP, 2.5ul RP, 2ul DNA, 5.5ul dH20.
- PSBIC3 (plasmid backbone - used as positive control): 12.5ul Taq readymix, 1ul FP, 1ul RP, 3ul DNA, 7.5ul dH20.
PCR:
We do not know yet whether this program was actually ran as it is written below, but here are the instructions.
- Initial D: 95C 30s
- 30 cycles of:
- 95C 15s
- 55C 30s
- 68C 50s
- Final extension 65C 5 mins
- Hold 4C ∞
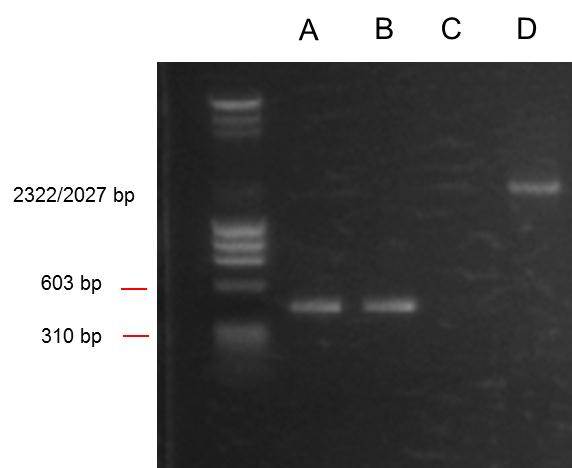
Visualisation and interpretation:
We loaded 3ul of each template with 2ul of leading buffer (5ul of ladder and 2ul of loading buffer), and electrophoresed on a 1% agarose gel at 76V for 40 mins.
From left:
- 1: 250-10000bp ladder (from hackspace). Ladder key
- 2: O. indofilex template from hackspace PCR product.
- 3: O. indofilex template from UCL PCR product.
- 4: ANF 1 PCR product.
- 5: ANF 2 PCR product.
- 6: ANF 3 PCR product.
- 7: PSB1C3 PCR product.
So we got no bands from the O. indofilex gDNA reactions, but we did get bands at the right position (285 bp) from 2/3 of the ANF template ones. ANF templates were ANF genes that had been previously PCRed with the non prefix/suffix primers. We also got a band from the positive control at the right position (2070bp).
Restriction digest and ligation
This step involves the insertion of the gene (ANF / MR in our case) into the plasmid backbone (PSB1C3). First the plasmid and the gene are incubated in separate reactions with restriction enzymes, to cut them at the appropriate parts so they can be ligated together.
Restriction digest and ligation 12/9
Restriction digest
Required mass of DNA (ie. plasmid or gene) in each reaction is 250ng, so quantities of PSB1C3 (50.9ng/ul conc.) and ANF (19.2ng/ul conc.) are calculated with this in mind. EcoR1 and PST1 are the restriction enzymes. BSA prevents adhesion of the enzyme(s) to reaction tubes and pipette surfaces, and stabilizes some proteins during incubation. NEB2 is a buffer for the restriction enzymes.
Plasmid backbone reaction (Total volume = 25ul)
- PSB1C - 4.9ul
- EcoR1 - 0.5ul
- PST1 - 0.5ul
- BSA - 0.3ul
- dH20 - 16.3ul
- NEB2 - 2.5ul
'ANF reaction (Total volume = 25ul)
- ANF - 13ul
- EcoR1 - 0.5ul
- PST1 - 0.5ul
- BSA - 0.3ul
- dH20 - 8.2ul
- NEB2 - 2.5ul
Control for ligation reaction (Total volume = 25ul)
- PSB1C - 4.9ul
- PST1 - 0.5ul
- BSA - 0.3ul
- dH20 - 16.8ul
- NEB2 - 2.5ul
Incubated at 37C 30mins, 80C 20mins.
Ligation
Ligation (Total volume = 20ul)
- Insert - 2ul
- Plasmid - 2ul
- 10xT4 buffer - 2ul
- T4 DNA ligase - 1ul
- dH20 - 13ul
Positive control (Total volume = 20ul)
- Plasmid control - 2ul
- 10xT4 buffer - 2ul
- T4 DNA ligase - 1ul
- dH20 - 15ul
Negative control' (Total volume = 20ul)
- Plasmid control - 2ul
- 10xT4 buffer - 2ul
- dH20 - 16ul
Incubated room temp 10mins, ligase heat inactivated at 80C for 20mins.
Transformation into E.coli W3110 cells
Miniprep: Plasmid DNA Extraction
TBA
Analytical digest
Characterisation
Unfortunately, there was no time to characterize (show it work) the biobrick. There is a simple plate assay that could have been used to test the activity of the protein produced. Protocol given by UCL: In order to express the gene, the biobrick we have now need to be cut with restriction enzymes whose restriction sites flank the genes to be ligated with a promoter and RBS (Ribosome binding site). It could be characterized by ligating (sticking) onto an expression vector (a plasmid that is commercially available that contains a promoter and ribosome binding site). After transformation do the following:
- Pick colony from ligation plates into 2ml LB. Incubate @37 until the OD reach approx 0.5.
- Perform serial dilutions of half of the liquid culture and plate
- Freeze the other half of the culture at -30C overnight.
- Next morning, perform the same serial dilutions on the remaining liquid culture and plate.
- Count the number of colonies on the plate before freezing and after freezing.