Project:Sex typing with amelogenin/results-20120212
Mailing list discussion
To think about
- DNA extraction: perhaps try several things, but especially:
- Detergent
- Salt
- DNA extraction: try centrifuging DNA
- DNA extraction: perhaps test by combining some extracted DNA with dilute EtBr solution.
- Primers: can we tell that they are there? Try combining some with EtBr and viewing.
- PCR: Do tests:
- Without primers or template
- Without template
- Without master mix (?)
- PCR: Follow Wikipedia's basic melt, anneal, extend temperatures and times.
Experiments to run:
Gel run 1
- PCR, no master mix, no template, just primers and 55 ul of buffer (Couldn't do this one, ran out of primers)
- PCR, no master mix, just template and primers and 5ul of buffer (Couldn't do this one, ran out of template and primers)
- PCR, master mix and primers, no template and 30 ul of buffer
- PCR, master mix, primers, and template (from top of tube)
- PCR, master mix, primers, and template (from bottom of tube)
- PCR, master mix (old one), primers, and template (from top of tube)
- PCR, master mix (old one), primers, and template (from bottom of tube)
DNA extraction
- 6ml of saline solution in mouth
- swish for 10 seconds
- spit back into tube
- centrifuge for 5 minutes
- pour off supernatant (liquid on top) into sink.
- suspend tube in boiling water for 10 minutes
- place on ice for 1 minute
- centrifuge for 30 seconds
- transfer 200ul of supernatant to fresh tube. This contains DNA. (Note I didn't do this step -- instead I took liquid from the top of the tube or the bottom of the tube, depending on the lane.)
Protocol
- 10 ul of primer (at a strength of 100 micromoles), x2 (so 10ul for each primer)
- 25 ul of fast-start PCR Taq readymix
- 30 ul of DNA.
- 50 ul of mineral oil on top
PCR
- D = 30 seconds at 98 degress
- A = 40 seconds at 55 degrees (I think)
- E = 60 seconds at 72 degrees
- Repeat 25 times.
- start: 14:22
- end: 16:02
Gel lanes (viewed with the gel oriented so the lanes are at the top)
- Nothing
- master mix and primers, no template and 30 ul of buffer
- master mix, primers, and template (from top of tube)
- master mix, primers, and template (from bottom of tube)
- Ladder, 6ul
- master mix (old one), primers, and template (from top of tube)
- master mix (old one), primers, and template (from bottom of tube)
- Nothing
Started electrophoresis at 16:11 at 100v.
First visualisation taken after ~20 minutes of electrophoresis.
Second visualisation taken after ~45 minutes of electrophoresis.
Results
All these images can be viewed in full resolution, but the thumbnails don't work because MediaWiki sucks.
- Fixed. Ms7821 23:34, 12 February 2012 (UTC)
- Much obliged! - NFD, 23:49, 2012-02-12
DNA was extracted following the protocol above: saline solution mouth-wash followed by 10 minutes of immersion in boiling water.
PCR and electrophoresis was performed using the above protocols.
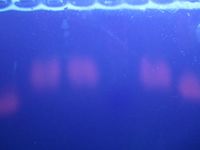
After 20 minutes, bands were observed in lanes 2, 3, 4, 6, and 7. The ladder was also visible in lane 5 but can't be seen in this photo.
After 45 minutes, the bands had become very blurry.
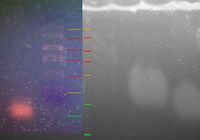
Here is a close-up of the middle portion of the gel. The ladder is faintly visible.
Looking for the ladder and its relation to the bands. On the left is the gel from last Wednesday. The lines in red are clearly lines in the ladder, the orange lines are putative ladder (less certain), and the green lines are the bands. If the interpretation presented here is correct, then this result repeats the results of last Wednesday.
Discussion
- The bands are very blurry and wonky, as if the agar was of varying densities throughout.
- Bands on lanes 3, 4, and 6 travelled less far than bands on lanes 2 and 7. Lane 2 did not include template (i.e. DNA); and I ran out of template in lane 7. So this is an encouraging result.
- The bands are within the bounds of the ladder this time.
- Bands on lanes 3, 4, and 6 are more blurry than bands on lanes 2 and 7. One exciting theory is that bands on those lanes actually worked, and the blurriness is because of the 200 base-pair difference between the two amelogenin sequences; whereas the bands on lanes 2 and 7 contain un-used primer and master mix, which is a) shorter sequences and b) more well-defined (presumably because there is less of a difference between the lengths).
Future
- Try to get the agar more consistent
- Electrophorese for 20 or 30 minutes, but no longer
- Chances are we *haven't* ruined our old master mix, which is great (£)
- We should aliquot our master mixes so we don't have to put them through any more thaw / freeze cycles.
- We may have too much template (surprisingly, this can be a problem)
- Do we need hot start?
Basically I think we are screwing up the PCR stage: using the wrong temperatures, too-short timings, or whatever. Here is a paper on sex typing with amelogenin:
http://library-resources.cqu.edu.au/JFS/PDF/vol_39/iss_6/JFS396941356.pdf
The paragraph relevant to PCR states:
Genomic DNA samples were amplified in a 100 µl reaction volume containing 0.2nM of each dNTP, 50 mM KCl, 10 mM Tris.HCl pH 8.3, 4.0 mM MgCl_2, 120 pmoles of each primer, and 2.5 U Taq polymerase. PCR was run for 30 cycles of 94 C for 1 min (denature), 65 C for 2 min (anneal), and 72 C for 3 min (extend). Following amplification, the PCR products were analyzed by electrophoresis on 1.2% agarose gels run with 1 X TBE (89 mM Tris borate, 89 mM boric acid, 2 mM EDTA) or 6% nonenaturing polyacrylamide gels run with 1 X TBE. The gels were stained with ethidium bromide and the results were visualized by fluorescence under ultraviolet light.
So, we need to:
- Run each PCR stage for longer (2 to 3 times as long)
- Use a higher annealing temperature